Examples#
Bellow you will find two model examples adopted from the Dendrify paper.
Example 1 | A basic compartmental model with passive dendrites
Example 2 | A reduced compartmental model capturing active dendritic properties
Tip
By clicking the “Open in Colab” button located under each example, you can run in your browser (without locally installing Dendrify or Brian) an interactive Jupyter notebook that reproduces the respective neuron models and simulation results.
Example 1 | A basic compartmental model with passive dendrites.
In this example we show that even rudimentary models can reproduce essentia neuronal properties such as the electrical segmentation caused by dendrites This allows multiple integration sites to coexist within a neuron and dendrite to operate semi-autonomously from the soma, while greatly affecting neuronal output.
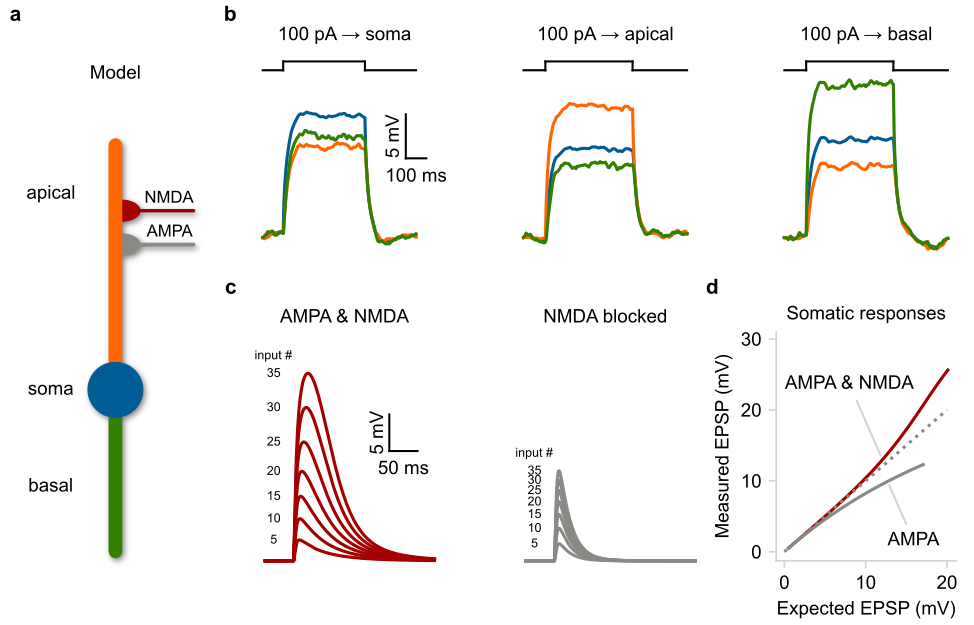
a) Schematic illustration of a compartmental model consisting of a soma (spiking unit) and two dendrites (passive integrators). The apical dendrite can integrate excitatory synapses comprising AMPA and NMDA currents. b) Membrane voltage responses to current injections of the same amplitude are applied individually to each compartment. Notice the electrical segregation caused by the resistance between the three neuronal compartments. c Somatic responses to a varying number of simultaneous synaptic inputs (5–35 synapses). Left: control EPSPs, Right: EPSPs in the presence of NMDA blockers. d) Input-output function of the apical dendrite as recorded at the soma. The dotted line represents a linear function. Notice the shift from supralinear to the sublinear mode when NMDARs are blocked.
Example 2: A reduced compartmental model capturing active dendritic properties.
In this example we show that reduced compartmental I&F models, equipped with event-driven dendritic spiking mechanisms can faithfully reproduce a broad range of dendritic properties such as: i) Supralinear input integration, ii) dendrite-specific spiking threshold, iii) distance-dependent filtering, iv) backpropagation of somatic spikes.
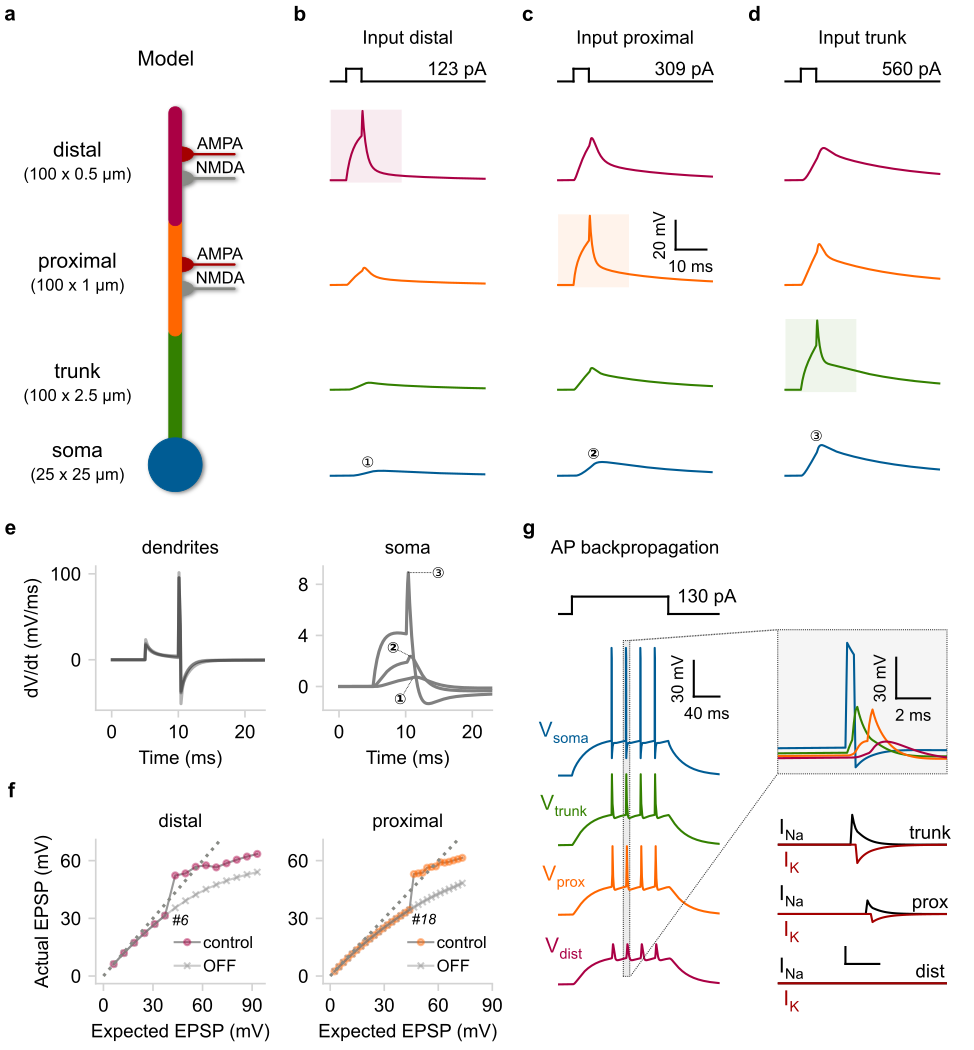
a) Schematic illustration of a compartmental model consisting of a soma (leaky I&F) and three dendritic segments (trunk, proximal, distal) equipped with Na+ VGICs. The distal and proximal segments can also receive AMPA and NMDA synapses. b–d) Rheobase current injections (5 ms square pulses) for dSpike generation were applied individually to each dendritic segment. Shaded areas: location of current injection and dSpike initiation. Top: stimulation protocol showing the current threshold for a single dSpike (rheobase current). e) First temporal derivative of dendritic (left) and somatic (right) voltage traces from panels (b–d). f) Input–output function of the distal (left) and proximal (right) segment as recorded from the corresponding dendritic locations. We also indicate the number of quasi-simultaneously activated synapses (ISI = 0.1 ms) needed to elicit a single dSpike in each case. OFF: deactivation of Na+ dSpikes. Dashed lines: linear input–output relationship. g) Left: Backpropagating dSpikes are generated in response to somatic current injections. The short-amplitude spikelets detected in the distal branch are subthreshold voltage responses for dSpike initiation. Right: Magnified and superimposed voltage traces (top) from the dashed box (left). Bottom: dendritic voltage-activated currents responsible for dSpikes generation in each dendritic segment.