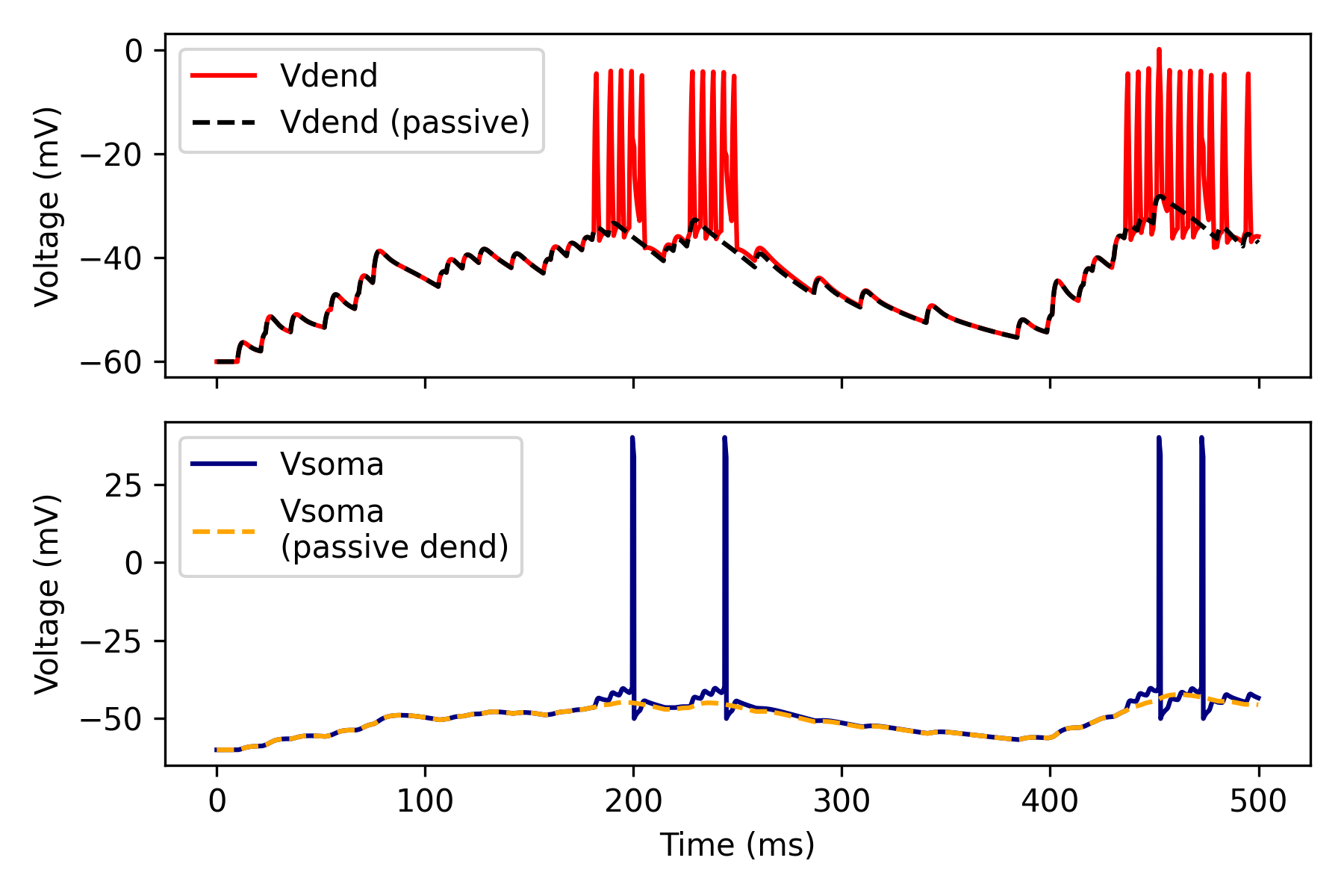
Active vs passive dendrites#
In pyramidal neurons, distal synapses have often a minute effect on the somatic membrane potential due to strong dendritic attenuation. However, the activation of dendritic spikes can amplify synaptic inputs that are temporally correlated, increasing the probability of somatic AP generation.
In this example we show:
How to create a compartmental model with passive or active dendrites.
How dendritic spiking may affect somatic AP generation.
import brian2 as b
from brian2.units import Hz, ms, mV, nS, pF
from dendrify import Dendrite, NeuronModel, Soma
b.prefs.codegen.target = 'numpy' # faster for simple simulations
# Create neuron model with passive dendrites
soma = Soma('soma', cm_abs=200*pF, gl_abs=10*nS)
dend = Dendrite('dend', cm_abs=50*pF, gl_abs=2.5*nS)
dend.synapse('AMPA', tag='x', g=3*nS, t_decay=2*ms)
dend.synapse('NMDA', tag='x', g=3*nS, t_decay=60*ms)
model_passive = NeuronModel([(soma, dend, 15*nS)], v_rest=-60*mV)
# Add dendritic spikes and create a neuron model with active dendrites
dend.dspikes('Na', g_rise=30*nS, g_fall=14*nS)
model_active = NeuronModel([(soma, dend, 15*nS)], v_rest=-60*mV)
model_active.config_dspikes(
'Na', threshold=-35*mV,
duration_rise=1.2*ms, duration_fall=2.4*ms,
offset_fall=0.2*ms, refractory=5*ms,
reversal_rise='E_Na', reversal_fall='E_K')
# Create a neuron group with passive dendrites
neuron_passive, reset_p = model_passive.make_neurongroup(
1, method='euler',
threshold='V_soma > -40*mV',
reset='V_soma = 40*mV',
second_reset='V_soma=-50*mV',
spike_width=0.8*ms,
refractory=4*ms)
# Create a neuron group with active dendrites
neuron_active, reset_a = model_active.make_neurongroup(
1, method='euler',
threshold='V_soma > -40*mV',
reset='V_soma = 40*mV',
second_reset='V_soma=-50*mV',
spike_width=0.8*ms,
refractory=4*ms)
# Create random Poisson input
Input_p = b.PoissonGroup(5, rates=20*Hz)
Input_a = b.PoissonGroup(5, rates=20*Hz)
# Create synapses
S_p = b.Synapses(Input_p, neuron_passive,
on_pre='s_AMPA_x_dend += 1; s_NMDA_x_dend += 1')
S_p.connect(p=1)
S_a = b.Synapses(Input_a, neuron_active,
on_pre='s_AMPA_x_dend += 1; s_NMDA_x_dend += 1')
S_a.connect(p=1)
# Record voltages
vars = ['V_soma', 'V_dend']
M_p = b.StateMonitor(neuron_passive, vars, record=True)
M_a = b.StateMonitor(neuron_active, vars, record=True)
# Run simulation
b.seed(123) # for reproducibility
net_passive = b.Network(neuron_passive, reset_p, Input_p, S_p, M_p)
net_passive.run(500*ms)
b.start_scope() # clear previous simulation
b.seed(123) # for reproducibility
net_active = b.Network(neuron_active, reset_a, Input_a, S_a, M_a)
net_active.run(500*ms)
# Visualize results
time_p = M_p.t/ms
vs_p = M_p.V_soma[0]/mV
vd_p = M_p.V_dend[0]/mV
time_a = M_a.t/ms
vs_a = M_a.V_soma[0]/mV
vd_a = M_a.V_dend[0]/mV
fig, axes = b.subplots(2, 1, figsize=(6, 4), sharex=True)
ax0, ax1 = axes
ax0.plot(time_a, vd_a, label='Vdend', c='red')
ax0.plot(time_p, vd_p, '--', label='Vdend (passive)', c='black')
ax0.set_ylabel('Voltage (mV)')
ax0.legend(loc=2)
ax1.plot(time_a, vs_a, label='Vsoma', c='navy')
ax1.plot(time_p, vs_p, '--', label='Vsoma\n(passive dend)', c='orange')
ax1.set_xlabel('Time (ms)')
ax1.set_ylabel('Voltage (mV)')
ax1.legend(loc=2)
fig.tight_layout()
b.show()