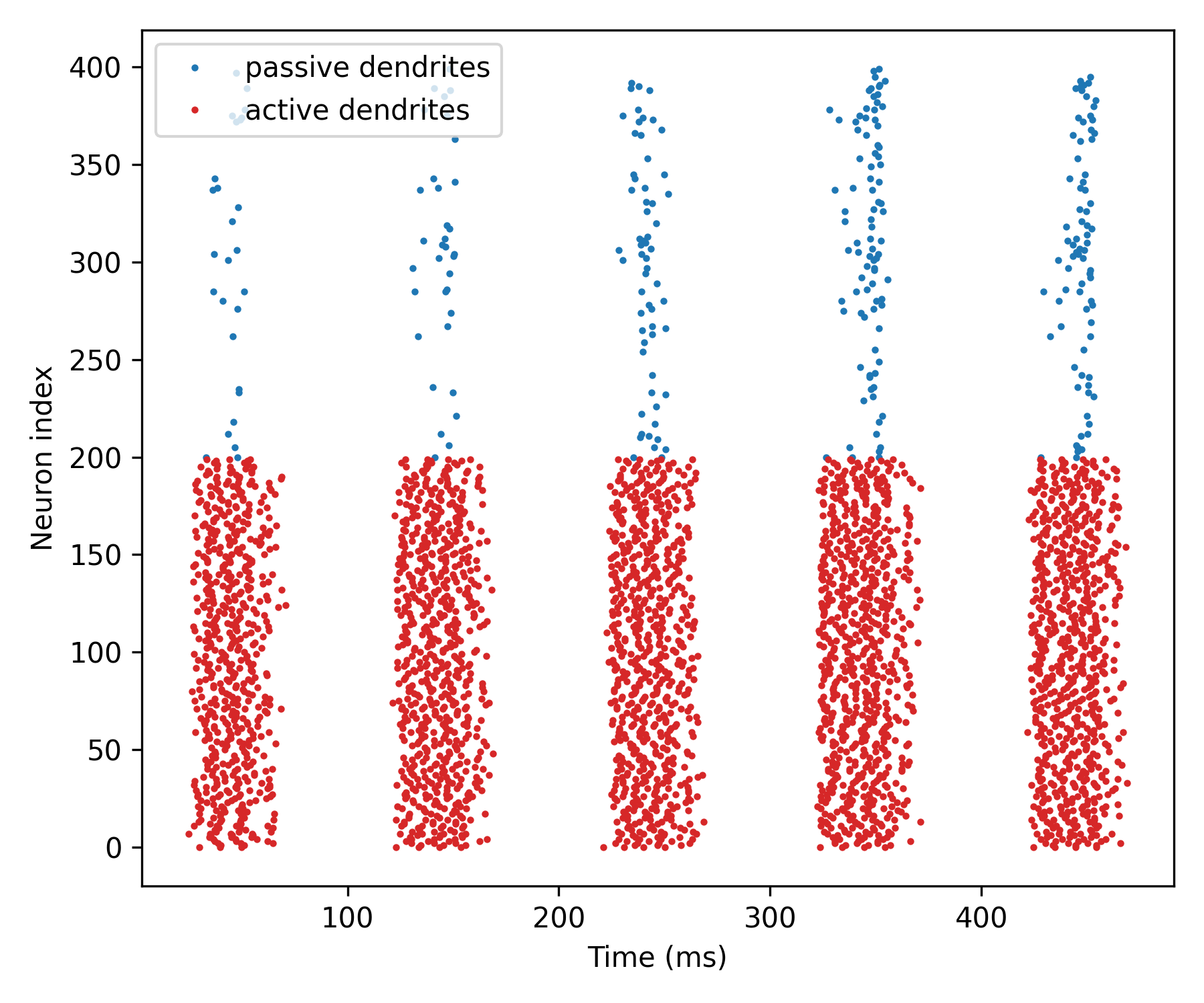
Networks of compartmental neurons#
In this example we show:
How to create a recurrent network of compartmental neurons.
How active dendrites can alter network responses.
import brian2 as b
from brian2.units import Hz, ms, mV, nS, pF
from dendrify import Dendrite, NeuronModel, Soma
b.prefs.codegen.target = 'numpy' # faster for simple simulations
# Create neuron model with passive dendrites
soma = Soma('soma', cm_abs=200*pF, gl_abs=10*nS)
dend = Dendrite('dend', cm_abs=50*pF, gl_abs=2.5*nS)
dend.synapse('AMPA', tag='external', g=1*nS, t_decay=5*ms)
dend.synapse('AMPA', tag='recurrent', g=1*nS, t_decay=5*ms)
model_passive = NeuronModel([(soma, dend, 15*nS)], v_rest=-60*mV)
# Add dendritic spikes and create a neuron model with active dendrites
dend.dspikes('Na', g_rise=30*nS, g_fall=14*nS)
model_active = NeuronModel([(soma, dend, 15*nS)], v_rest=-60*mV)
model_active.config_dspikes(
'Na', threshold=-35*mV,
duration_rise=1.2*ms, duration_fall=2.4*ms,
offset_fall=0.2*ms, refractory=5*ms,
reversal_rise='E_Na', reversal_fall='E_K')
# Create a neuron group with passive dendrites
neuron_passive, reset_p = model_passive.make_neurongroup(
200, method='euler',
threshold='V_soma > -40*mV',
reset='V_soma = 40*mV',
second_reset='V_soma=-50*mV',
spike_width=0.8*ms,
refractory=4*ms)
# Create a neuron group with active dendrites
neuron_active, reset_a = model_active.make_neurongroup(
200, method='euler',
threshold='V_soma > -40*mV',
reset='V_soma = 40*mV',
second_reset='V_soma=-50*mV',
spike_width=0.8*ms,
refractory=4*ms)
# Create random Poisson input
# Protocol: five 50 ms blocks of 50 Hz stimulation, followed by 50 ms of silence
stimulus = b.TimedArray(b.tile([50., 0.], 5)*Hz, dt=50.*ms)
Input_a = b.PoissonGroup(200, rates='stimulus(t)')
Input_p = b.PoissonGroup(200, rates='stimulus(t)')
# Create synapses for external input
S_input_passive = b.Synapses(
Input_p, neuron_passive,
on_pre='s_AMPA_external_dend += 1')
S_input_passive.connect(p=0.2)
S_input_active = b.Synapses(
Input_a, neuron_active,
on_pre='s_AMPA_external_dend += 1')
S_input_active.connect(p=0.2)
# Create recurrent synapses
S_recurrent_passive = b.Synapses(
neuron_passive, neuron_passive,
on_pre='s_AMPA_recurrent_dend += 1')
S_recurrent_passive.connect(p=0.1)
S_recurrent_active = b.Synapses(
neuron_active, neuron_active,
on_pre='s_AMPA_recurrent_dend += 1')
S_recurrent_active.connect(p=0.1)
# Record spikes
spikes_passive = b.SpikeMonitor(neuron_passive)
spikes_active = b.SpikeMonitor(neuron_active)
# Run simulation
b.seed(123) # for reproducibility
net_passive = b.Network(neuron_passive, reset_p, Input_p,
S_input_passive, S_recurrent_passive,
spikes_passive)
net_passive.run(550*ms)
b.start_scope() # clear previous simulation
b.seed(123) # for reproducibility
net_active = b.Network(neuron_active, reset_a, Input_a,
S_input_active, S_recurrent_active,
spikes_active)
net_active.run(550*ms)
# Visualize results
b.figure(figsize=[6, 5])
b.plot(spikes_passive.t/ms, spikes_passive.i + 200,
'.', ms=3, label='passive dendrites')
b.plot(spikes_active.t/ms, spikes_active.i,
'.', ms=3, c='C3', label='active dendrites')
b.xlabel('Time (ms)')
b.ylabel('Neuron index')
b.legend()
b.tight_layout()
b.show()