AdEx network + synapses#
The Dendrify implementation of the Adaptive exponential integrate-and-fire model (adapted from Brian’s examples).
In this example, we also explore:
How to add random Poisson synaptic input.
How to create a basic network model.
Resources:
import brian2 as b
from brian2.units import Hz, ms, mV, nA, nS, pF
from dendrify import PointNeuronModel
b.prefs.codegen.target = 'numpy' # faster for simple simulations
b.seed(1234) # for reproducibility
# Create neuron model and add AMPA equations
model = PointNeuronModel(model='adex',
cm_abs=281*pF,
gl_abs=30*nS,
v_rest=-70.6*mV)
model.synapse('AMPA', tag='x', g=0.5*nS, t_decay=2*ms)
model.synapse('NMDA', tag='x', g=0.5*nS, t_decay=50*ms)
# Include adex parameters
model.add_params({'Vth': -50.4*mV,
'DeltaT': 2*mV,
'tauw': 144*ms,
'a': 4*nS,
'b': 0.0805*nA,
'Vr': -70.6*mV})
# Create a NeuronGroup
neuron = model.make_neurongroup(N=100, threshold='V>Vth+5*DeltaT',
reset='V=Vr; w+=b',
method='euler')
# Create a Poisson input
Input = b.PoissonGroup(200, rates=100*Hz)
# Randomly connect Poisson input to NeuronGroup
S = b.Synapses(Input, neuron, on_pre='s_AMPA_x += 1; s_NMDA_x += 1')
S.connect(p=0.25)
# Record voltages and spike times
trace = b.StateMonitor(neuron, 'V', record=0)
spikes = b.SpikeMonitor(neuron)
# Run simulation
b.run(200 * ms)
# Trick to draw nicer spikes in I&F models
vm = trace[0].V[:]
for t in spikes.spike_trains()[0]:
i = int(t / b.defaultclock.dt)
vm[i] = 20*mV
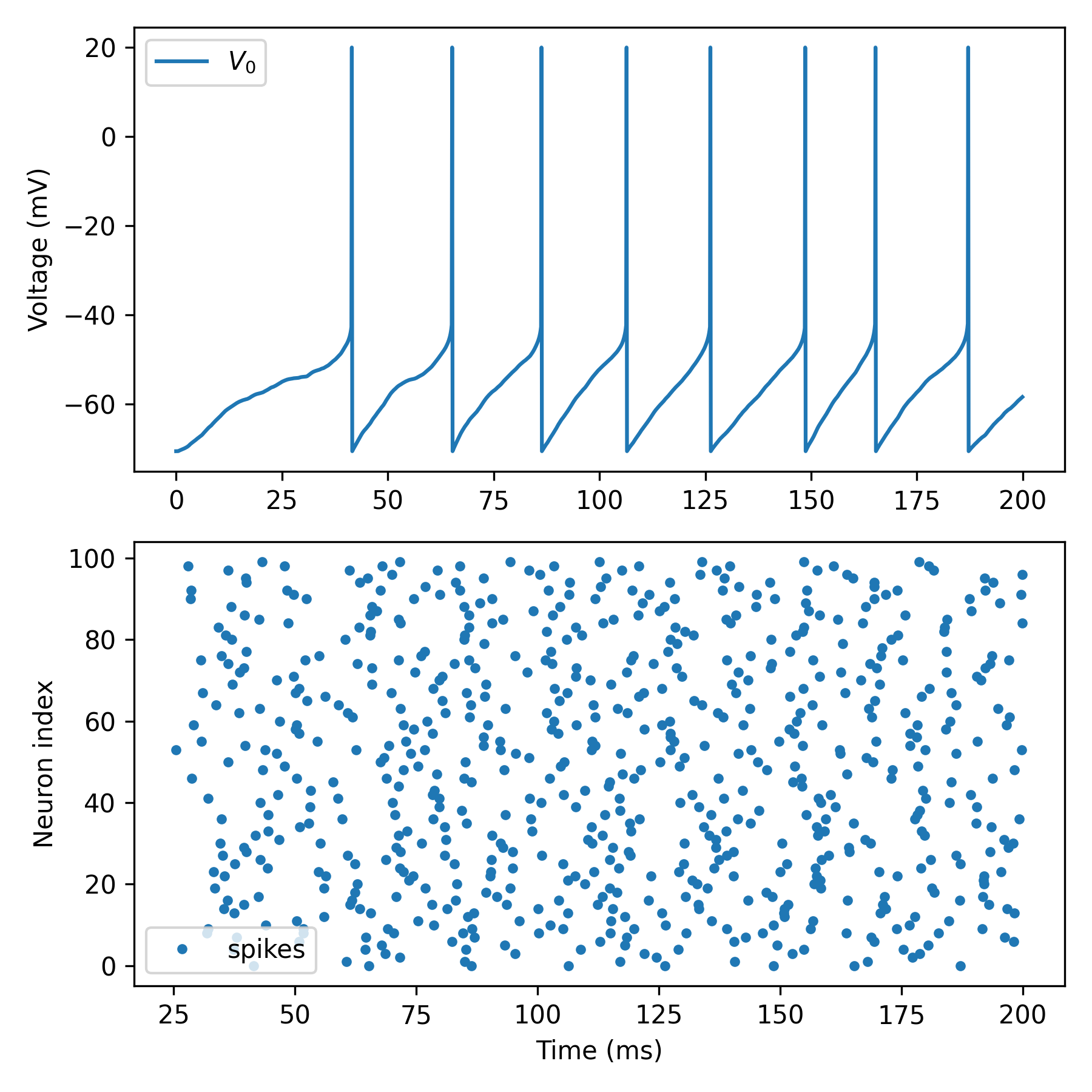
# Plot results
fig, axes = b.subplots(2, 1, figsize=[6, 6])
ax1, ax2 = axes
ax1.plot(trace.t / ms, vm / mV, label='$V_0$')
ax1.set_ylabel('Voltage (mV)')
ax1.legend()
ax2.plot(spikes.t/ms, spikes.i, '.', label='spikes')
ax2.set_xlabel('Time (ms)')
ax2.set_ylabel('Neuron index')
ax2.legend()
fig.tight_layout()
b.show()