NMDA synapses#
NMDA receptors, like AMPA receptors, are ionotropic glutamate receptors. They are responsible for excitatory synaptic transmission and have slower kinetics than AMPA receptors. NMDA receptors are voltage-dependent and require the postsynaptic membrane to be depolarized to remove the magnesium block and allow the influx of sodium and calcium ions.
In this example we show:
How to add and activate NMDA synapses in a few-compartmental model.
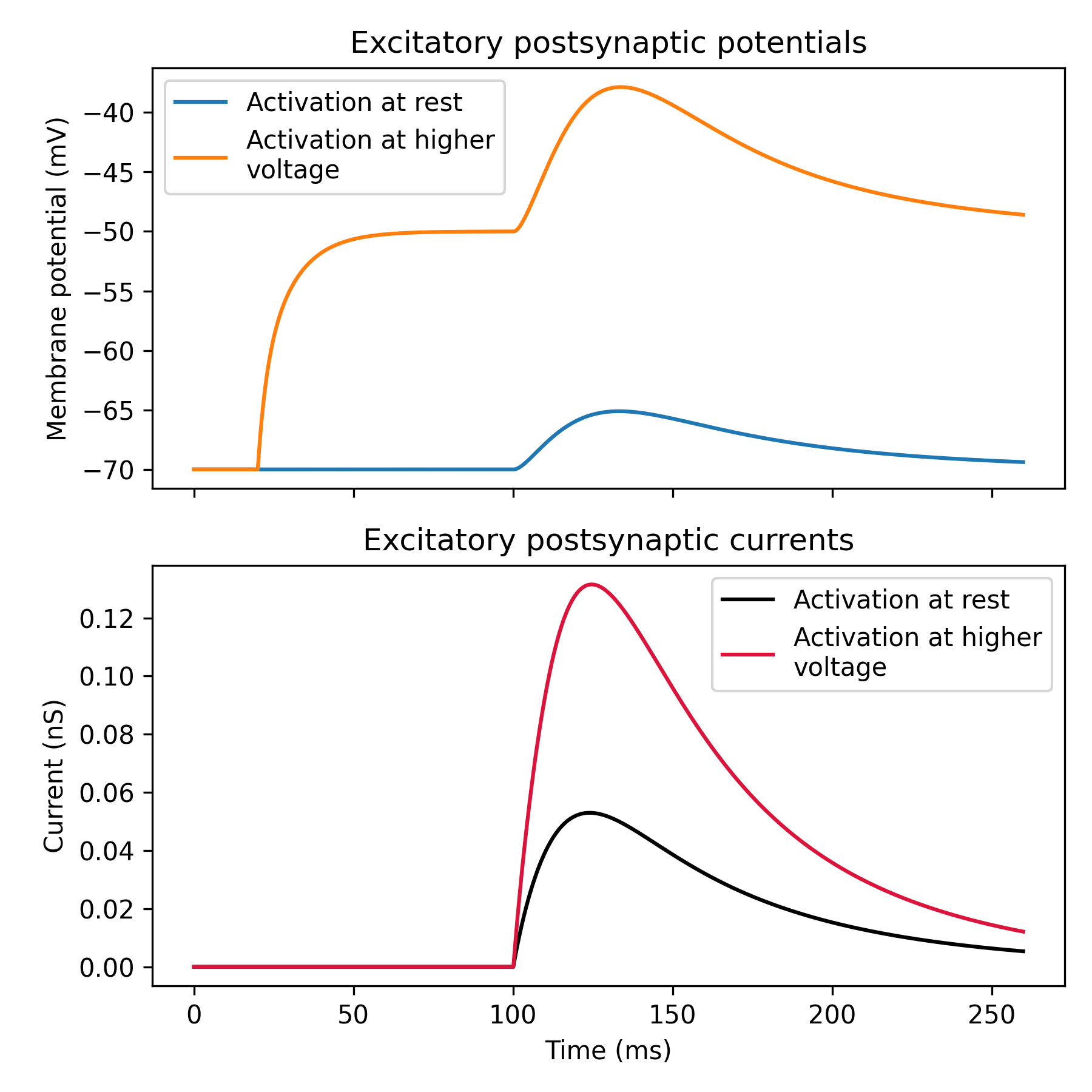
How NMDA synapses behave when activated at different membrane voltages.
import brian2 as b
from brian2.units import ms, mV, nS, pA, pF
from dendrify import Dendrite, NeuronModel, Soma
b.prefs.codegen.target = 'numpy' # faster for simple simulations
# Create a simple 3-compartment neuron model
soma = Soma('soma', cm_abs=100*pF, gl_abs=10*nS)
dend = Dendrite('dend', cm_abs=50*pF, gl_abs=5*nS)
dend.synapse('NMDA', tag='foo', g=2*nS, t_rise=10*ms, t_decay=60*ms)
# Merge the compartments into a single neuron model
model = NeuronModel([(soma, dend, 10*nS)], v_rest=-70*mV)
# Create 2 neurons (no somatic spiking for simplicity)
neurons = model.make_neurongroup(2, method='euler')
# Each neuron receives a spike at t=10ms at different dendrites
input_spikes = b.SpikeGeneratorGroup(2, [0, 1], [100, 100]*ms)
slow_ampa = b.Synapses(input_spikes, neurons, on_pre='s_NMDA_foo_dend += 1')
slow_ampa.connect(i=0, j=0)
fast_ampa = b.Synapses(input_spikes, neurons, on_pre='s_NMDA_foo_dend += 1')
fast_ampa.connect(i=1, j=1)
# Create monitors
variables = ['V_dend', 'I_NMDA_foo_dend']
M = b.StateMonitor(neurons, variables, record=True)
# Run the simulation
b.run(20*ms)
neurons[1].I_ext_dend = 200*pA
b.run(240*ms)
# Visualize results
time = M.t/ms
vd1, vd2 = M.V_dend[0]/mV, M.V_dend[1]/mV
nmda1, nmda2 = M.I_NMDA_foo_dend[0]/nS, M.I_NMDA_foo_dend[1]/nS
fig, axes = b.subplots(2, 1, sharex=True, figsize=(6, 6))
ax0, ax1 = axes
ax0.set_title('Excitatory postsynaptic potentials')
ax0.plot(time, vd1, label='Activation at rest')
ax0.plot(time, vd2, label='Activation at higher\nvoltage')
ax0.set_ylabel('Membrane potential (mV)')
ax0.legend()
ax1.set_title('Excitatory postsynaptic currents')
ax1.plot(time, nmda1, c='black', label='Activation at rest')
ax1.plot(time, nmda2, c='crimson', label='Activation at higher\nvoltage')
ax1.set_xlabel('Time (ms)')
ax1.set_ylabel('Current (nS)')
ax1.legend()
fig.tight_layout()
b.show()